Parameter Selection for HDBSCAN*¶
While the HDBSCAN class has a large number of parameters that can be set on initialization, in practice there are a very small number of parameters that have significant practical effect on clustering. We will consider those major parameters, and consider how one may go about choosing them effectively.
Selecting min_cluster_size¶
The primary parameter to effect the resulting clustering is
min_cluster_size. Ideally this is a relatively intuitive parameter
to select – set it to the smallest size grouping that you wish to
consider a cluster. It can have slightly non-obvious effects however.
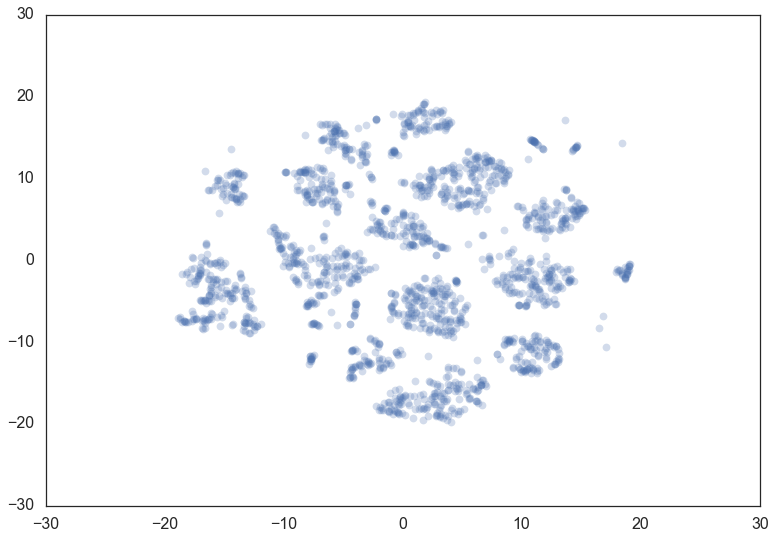
Let’s consider the digits dataset from sklearn. We can project the data
into two dimensions to visualize it via t-SNE.
digits = datasets.load_digits()
data = digits.data
projection = TSNE().fit_transform(data)
plt.scatter(*projection.T, **plot_kwds)
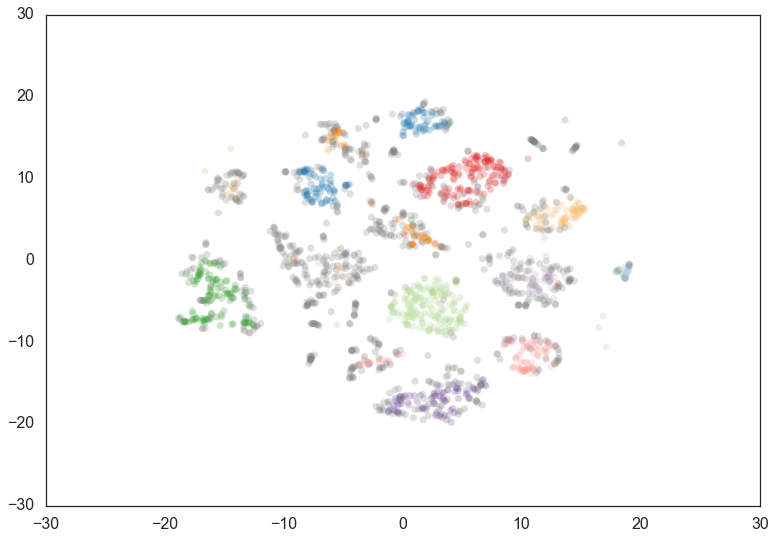
If we cluster this data in the full 64 dimensional space with HDBSCAN* we
can see some effects from varying the min_cluster_size.
We start with a min_cluster_size of 15.
clusterer = hdbscan.HDBSCAN(min_cluster_size=15).fit(data)
color_palette = sns.color_palette('Paired', 12)
cluster_colors = [color_palette[x] if x >= 0
else (0.5, 0.5, 0.5)
for x in clusterer.labels_]
cluster_member_colors = [sns.desaturate(x, p) for x, p in
zip(cluster_colors, clusterer.probabilities_)]
plt.scatter(*projection.T, s=50, linewidth=0, c=cluster_member_colors, alpha=0.25)
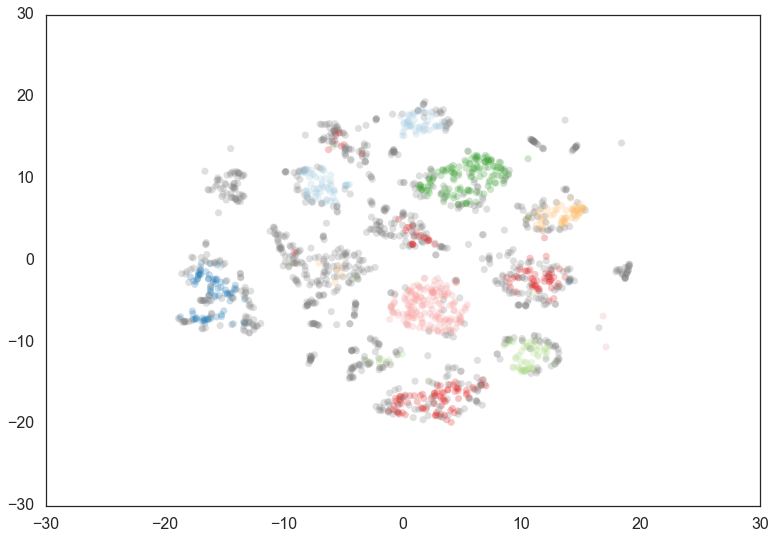
Increasing the min_cluster_size to 30 reduces the number of
clusters, merging some together. This is a result of HDBSCAN*
reoptimizing which flat clustering provides greater stability under a
slightly different notion of what constitutes a cluster.
clusterer = hdbscan.HDBSCAN(min_cluster_size=30).fit(data)
color_palette = sns.color_palette('Paired', 12)
cluster_colors = [color_palette[x] if x >= 0
else (0.5, 0.5, 0.5)
for x in clusterer.labels_]
cluster_member_colors = [sns.desaturate(x, p) for x, p in
zip(cluster_colors, clusterer.probabilities_)]
plt.scatter(*projection.T, s=50, linewidth=0, c=cluster_member_colors, alpha=0.25)
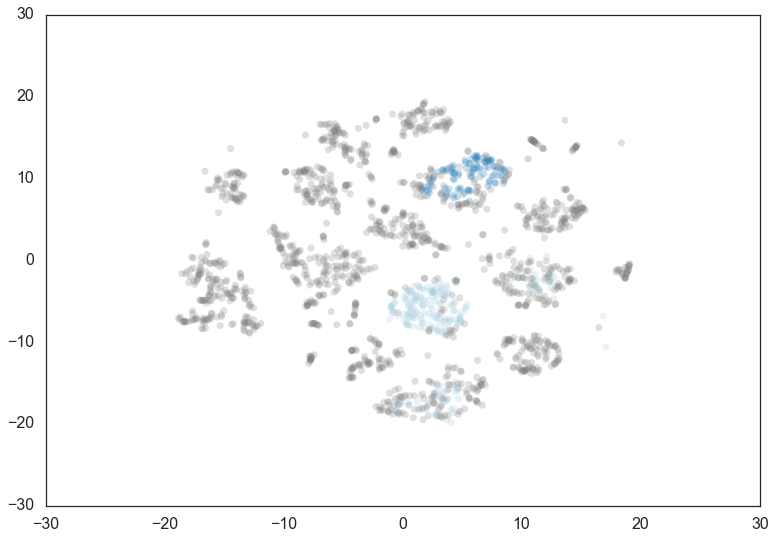
Doubling the min_cluster_size again to 60 gives us just two clusters
– the really core clusters. This is somewhat as expected, but surely
some of the other clusters that we had previously had more than 60
members? Why are they being considered noise? The answer is that
HDBSCAN* has a second parameter min_samples. The implementation
defaults this value (if it is unspecified) to whatever
min_cluster_size is set to. We can recover some of our original
clusters by explicitly providing min_samples at the original value
of 15.
clusterer = hdbscan.HDBSCAN(min_cluster_size=60).fit(data)
color_palette = sns.color_palette('Paired', 12)
cluster_colors = [color_palette[x] if x >= 0
else (0.5, 0.5, 0.5)
for x in clusterer.labels_]
cluster_member_colors = [sns.desaturate(x, p) for x, p in
zip(cluster_colors, clusterer.probabilities_)]
plt.scatter(*projection.T, s=50, linewidth=0, c=cluster_member_colors, alpha=0.25)
clusterer = hdbscan.HDBSCAN(min_cluster_size=60, min_samples=15).fit(data)
color_palette = sns.color_palette('Paired', 12)
cluster_colors = [color_palette[x] if x >= 0
else (0.5, 0.5, 0.5)
for x in clusterer.labels_]
cluster_member_colors = [sns.desaturate(x, p) for x, p in
zip(cluster_colors, clusterer.probabilities_)]
plt.scatter(*projection.T, s=50, linewidth=0, c=cluster_member_colors, alpha=0.25)
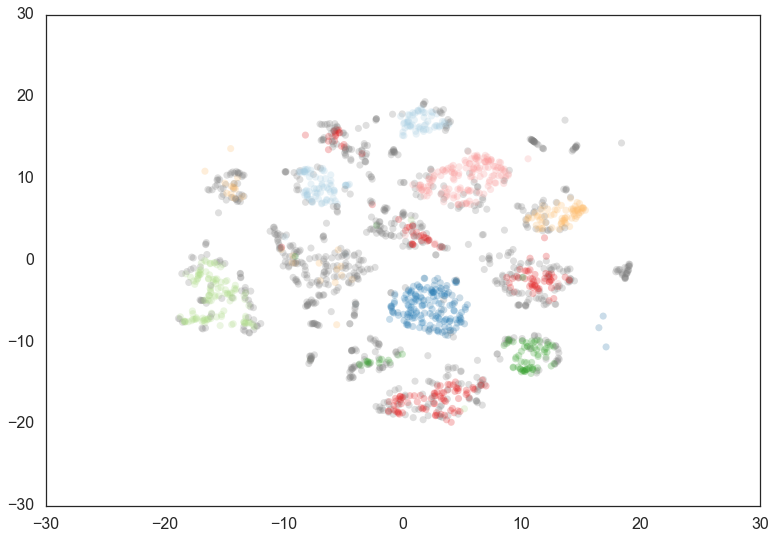
As you can see this results in us recovering something much closer to
our original clustering, only now with some of the smaller clusters
pruned out. Thus min_cluster_size does behave more closely to our
intuitions, but only if we fix min_samples.
If you wish to explore different
min_cluster_sizesettings with a fixedmin_samplesvalue, especially for larger dataset sizes, you can cache the hard computation, and recompute only the relatively cheap flat cluster extraction using thememoryparameter, which makes use of joblib
Selecting min_samples¶
Since we have seen that min_samples clearly has a dramatic effect on
clustering, the question becomes: how do we select this parameter? The
simplest intuition for what min_samples does is provide a measure of
how conservative you want your clustering to be. The larger the value of
min_samples you provide, the more conservative the clustering –
more points will be declared as noise, and clusters will be restricted
to progressively more dense areas. We can see this in practice by
leaving the min_cluster_size at 60, but reducing min_samples to
1.
Note: adjusting
min_sampleswill result in recomputing the hard comptuation of the single linkage tree.
clusterer = hdbscan.HDBSCAN(min_cluster_size=60, min_samples=1).fit(data)
color_palette = sns.color_palette('Paired', 12)
cluster_colors = [color_palette[x] if x >= 0
else (0.5, 0.5, 0.5)
for x in clusterer.labels_]
cluster_member_colors = [sns.desaturate(x, p) for x, p in
zip(cluster_colors, clusterer.probabilities_)]
plt.scatter(*projection.T, s=50, linewidth=0, c=cluster_member_colors, alpha=0.25)
<matplotlib.collections.PathCollection at 0x120978438>
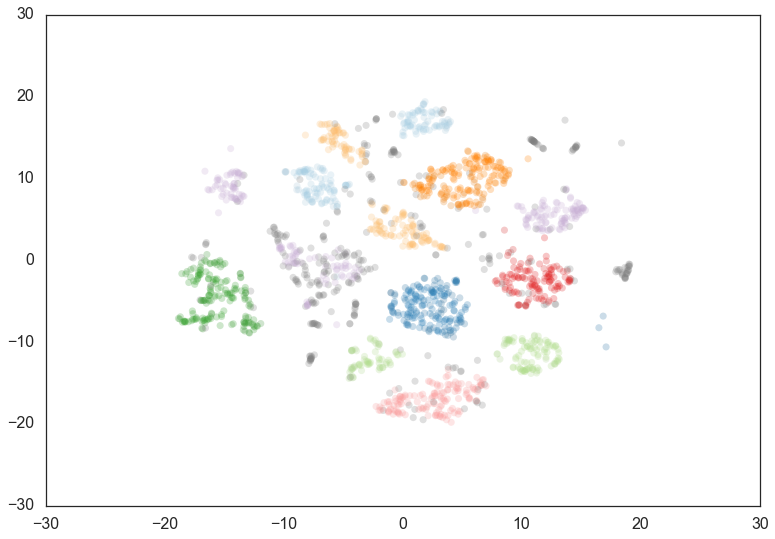
Now most points are clustered, and there are much fewer noise points.
Steadily increasing min_samples will, as we saw in the examples
above, make the clustering progressively more conservative, culminating
in the example above where min_samples was set to 60 and we had only
two clusters with most points declared as noise.
Selecting cluster_selection_epsilon¶
In some cases, we want to choose a small min_cluster_size because even groups of few points might be of interest to us.
However, if our data set also contains partitions with high concentrations of objects, this parameter setting can result in
a large number of micro-clusters. Selecting a value for cluster_selection_epsilon helps us to merge clusters in these regions.
Or in other words, it ensures that clusters below the given threshold are not split up any further.
The choice of cluster_selection_epsilon depends on the given distances between your data points. For example, set the value to 0.5 if you don’t want to
separate clusters that are less than 0.5 units apart. This will basically extract DBSCAN* clusters for epsilon = 0.5 from the condensed cluster tree, but leave
HDBSCAN* clusters that emerged at distances greater than 0.5 untouched. See Combining HDBSCAN* with DBSCAN for a more detailed demonstration of the effect this parameter
has on the resulting clustering.
Selecting alpha¶
A further parameter that effects the resulting clustering is alpha.
In practice it is best not to mess with this parameter – ultimately it
is part of the RobustSingleLinkage code, but flows naturally into
HDBSCAN*. If, for some reason, min_samples or cluster_selection_epsilon is not providing you
what you need, stop, rethink things, and try again with min_samples or cluster_selection_epsilon.
If you still need to play with another parameter (and you shouldn’t),
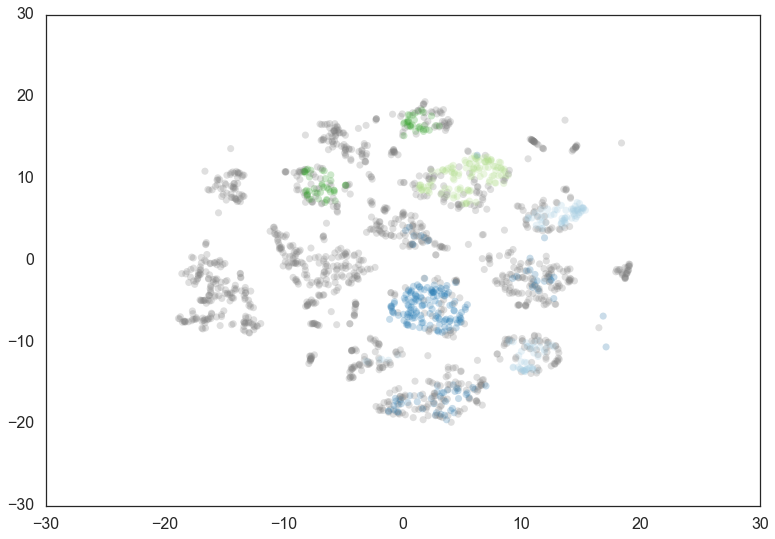
then you can try setting alpha. The alpha parameter provides a
slightly different approach to determining how conservative the
clustering is. By default alpha is set to 1.0. Increasing alpha
will make the clustering more conservative, but on a much tighter scale,
as we can see by setting alpha to 1.3.
Note: adjusting
alphawill result in recomputing the hard comptuation of the single linkage tree.
clusterer = hdbscan.HDBSCAN(min_cluster_size=60, min_samples=15, alpha=1.3).fit(data)
color_palette = sns.color_palette('Paired', 12)
cluster_colors = [color_palette[x] if x >= 0
else (0.5, 0.5, 0.5)
for x in clusterer.labels_]
cluster_member_colors = [sns.desaturate(x, p) for x, p in
zip(cluster_colors, clusterer.probabilities_)]
plt.scatter(*projection.T, s=50, linewidth=0, c=cluster_member_colors, alpha=0.25)
Leaf clustering¶
HDBSCAN supports an extra parameter cluster_selection_method to determine
how it selects flat clusters from the cluster tree hierarchy. The default
method is 'eom' for Excess of Mass, the algorithm described in
How HDBSCAN Works. This is not always the most desireable approach to
cluster selection. If you are more interested in having small homogeneous
clusters then you may find Excess of Mass has a tendency to pick one or two
large clusters and then a number of small extra clusters. In this situation
you may be tempted to recluster just the data in the single large cluster.
Instead, a better option is to select 'leaf' as a cluster selection
method. This will select leaf nodes from the tree, producing many small
homogeneous clusters. Note that you can still get variable density clusters
via this method, and it is also still possible to get large clusters, but
there will be a tendency to produce a more fine grained clustering than
Excess of Mass can provide.
Allowing a single cluster¶
In contrast, if you are getting lots of small clusters, but believe there
should be some larger scale structure (or the possibility of no structure),
consider the allow_single_cluster option. By default HDBSCAN* does not
allow a single cluster to be returned – this is due to how the Excess of
Mass algorithm works, and a bias towards the root cluster that may occur. You
can override this behaviour and see what clustering would look like if you
allow a single cluster to be returned. This can alleviate issue caused by
there only being a single large cluster, or by data that is essentially just
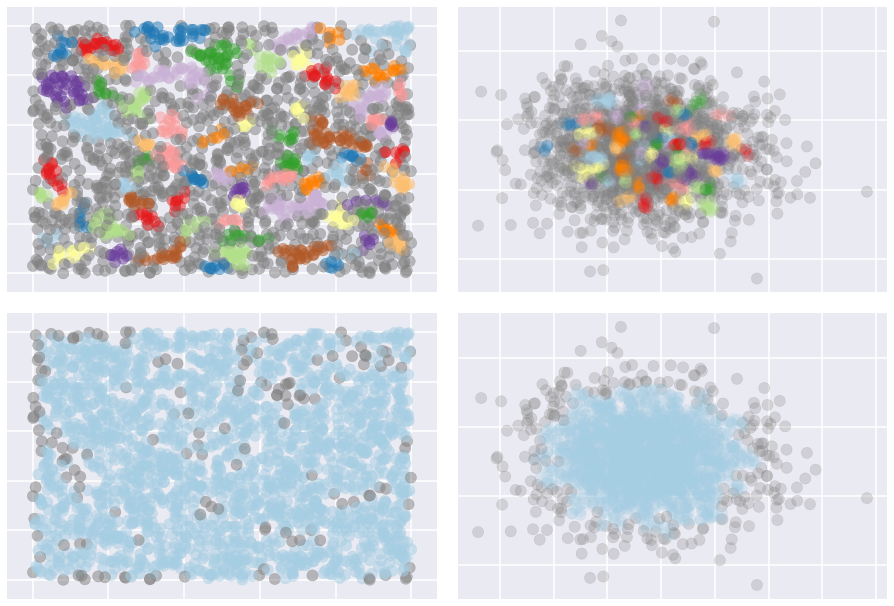
noise. For example, the image below shows the effects of setting
allow_single_cluster=True in the bottom row, compared to the top row
which used default settings.